Pulsed-field gel electrophoresis (PFGE) is a highly discriminative molecular typing technique that is used in epidemiological studies worldwide. PFGE is based upon the variable migration of large DNA restriction fragments in an electrical field of alternating polarity. By comparing the fingerprints of any two isolates, one can investigate if they belong to the same strain (i.e. the two isolates are clonal) or if they are genetically unrelated. The implementation of PFGE in international surveillance networks, such as PulseNet, and the availability of standardized protocols for important foodborne pathogens (E. coli, Listeria, Campylobacter, …) have facilitated the adoption of this technique. Although more recent typing methods are available, PFGE is still the gold standard in many national and international surveillance programs.
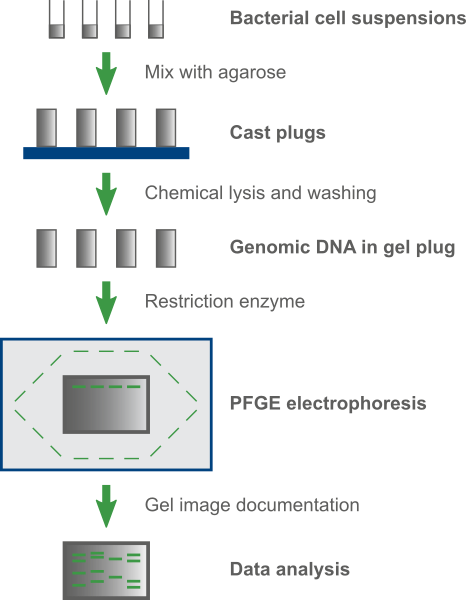
In practice, standardized cell suspensions of investigated isolates are embedded in agar to enhance DNA stability during the procedure. Then, bacterial cells are lysed and large fragments of the bacterial chromosome are generated by macro-restriction analysis. DNA fragments are separated during electrophoresis using an electrical field of alternating polarity, after which the fingerprints are visualized and documented.
PFGE analysis in BIONUMERICS
The BIONUMERICS software offers an integrated platform for the analysis of PFGE fingerprints. BIONUMERICS uses industry leading database engines that allow you to store all your epidemiological info and gel images in one database. Convenient wizards help you to define new fingerprint types, choose the optimal settings for normalization, resolution, background subtraction, smoothing, and band finding. The entire gel image pre-processing is contained in a powerful tab-based window, allowing easy access to re-edit the processing at any stage without losing any editing in another step.
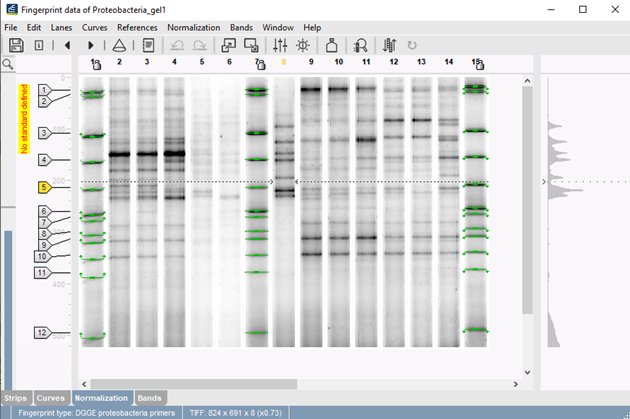
Our unique concept of reference systems makes it possible to automatically and reliably remap experiments run under different conditions. This important feature makes it possible for labs to exchange and compare electrophoresis data obtained with different conditions or setups.
Link fingerprint data to isolates in the BIONUMERICS database by easy drag-and-drop.
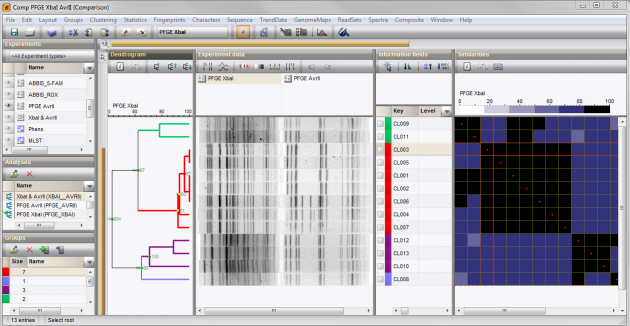
Create comparisons in a few mouse clicks: a wizard helps you to calculate similarity matrices and dendrograms using BIONUMERICS' rich set of statistical tools:
- Included band-based similarity coefficients: Jaccard, Dice, Jeffrey's X, Ochiai, Number of different bands.
- Choose between various cluster analysis methods: UPGMA, Ward, Neighbor Joining, Single Linkage, Complete Linkage, Create Graph.
Assess the significance of created clusters by calculating error flags and cophenetic correlations.
Set up identification projects starting from a set of reference strains.
