 The BIONUMERICS HIV drug resistance analysis application is a plugin-based expert system for automated prediction of antiretroviral drug resistance/susceptibility of HIV strains based on pol gene sequencing. It offers an open expert system that allows researchers to import and compare publicly available rule sets (algorithms) and create, adapt and (re)evaluate their own expert rules. The system does not just show resistance/susceptibility results, but reports the particular rules that are triggered and the mutations that are responsible for the triggering. The application offers a fully integrated and automated workflow starting from sequence assembly to final resistance reporting and is designed for professional high-throughput diagnostics.
The BIONUMERICS HIV drug resistance analysis application is a plugin-based expert system for automated prediction of antiretroviral drug resistance/susceptibility of HIV strains based on pol gene sequencing. It offers an open expert system that allows researchers to import and compare publicly available rule sets (algorithms) and create, adapt and (re)evaluate their own expert rules. The system does not just show resistance/susceptibility results, but reports the particular rules that are triggered and the mutations that are responsible for the triggering. The application offers a fully integrated and automated workflow starting from sequence assembly to final resistance reporting and is designed for professional high-throughput diagnostics.
A description of the background of the expert rule system is given in a poster presentation "An advanced expert rule system for HIV drug resistance prediction in the BIONUMERICS software".
Key features
The plugin offers a number of advantages over any other HIV drug resistance expert rule system:
- Full automatic batch assembly of HIV-1 pol gene sequences with detailed error and problem reporting and feedback. See poster Screening for influenza A H5N1 haemagglutinin and neuramidase mutations using the new Alignment & Mutation Analysis tool in the BIONUMERICS software for a description of the free batch sequence assembly plugin.
- Fully automated batch processing of assembled sequences, including alignment to wild-type (consensus) at nucleotide and amino acid level, detecting mutations and reporting problems and/or inconsistencies.
- Up-to-date prediction based on the latest releases of publicly available algorithm databases (HIVDB, ANRS, REGA and others), supporting score-based and S-I-R mapping algorithms.
- Detailed batch reporting of resistance levels for all currently known antiviral drugs.
- Expert rule system based on a unique Decision Network concept, allowing full monitoring for each sequence so that each particular mutation (or combination) leading to increased or decreased resistance can be identified.
- Combined reports comparing the results for multiple algorithms in detail.
- Decision Network concept allows you to edit, add and remove rules in an intuitive graphical environment and save modified algorithms under a different name.
- One integrated environment: possible to check sequence chromatograms for critical mutations or doubtful positions.
- All processing and analysis steps happen fully automatically on batches of any number of sequences and generate detailed reports, making the system extremely suitable for high-throughput screening.
- Powerful multi-user database and multi-experiment platform allows for the repository and management of information at all levels, ranging from patients to strains and samples.
Example screen views
Figure 1: Stored report of automated batch mutation screening, indicating frame shifts and degenerated positions on nucleotide sequences.
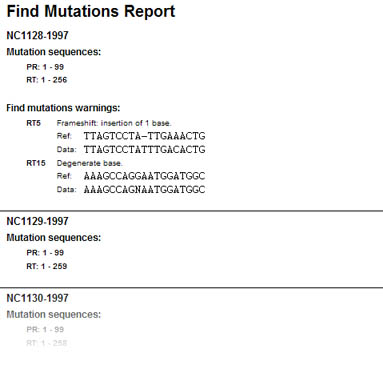
Figure 2: Evaluate resistance rules for available algorithms using the Decision Network.
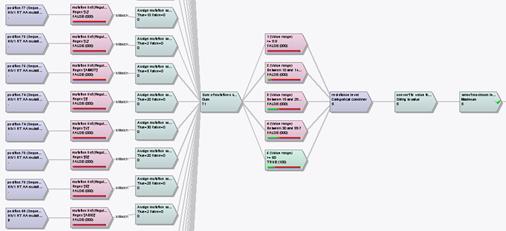
Figure 3: Concise overview of resistance patterns for multiple HIV strains; click on any strain or drug to display detailed report.
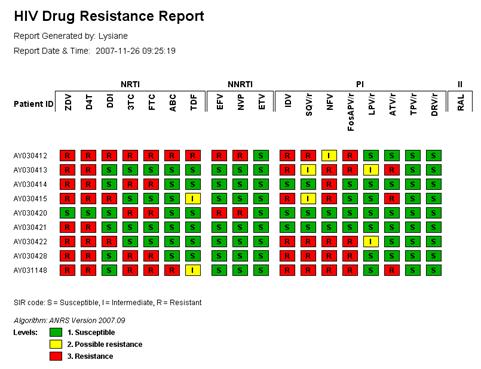
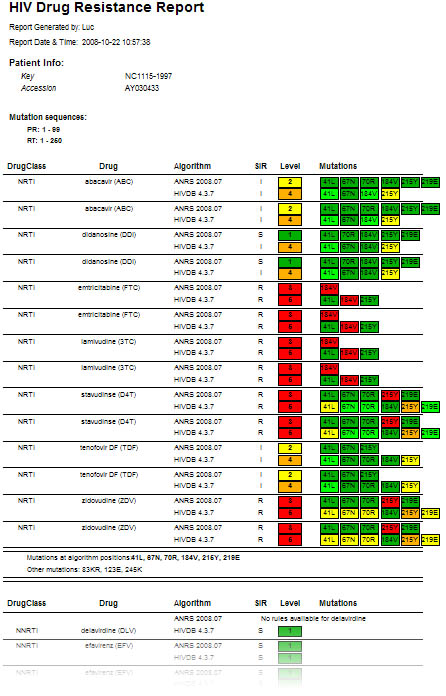
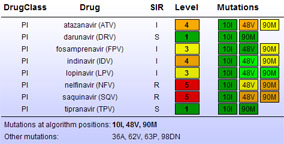
Figure 4: Detailed HIV strain resistance pattern report, indicating for each drug the individual mutations that lead to increased (yellow to red) or that have no influence on resistance (green) according to the algorithm used. Click on a drug to display the rule evaluation in the decision network.
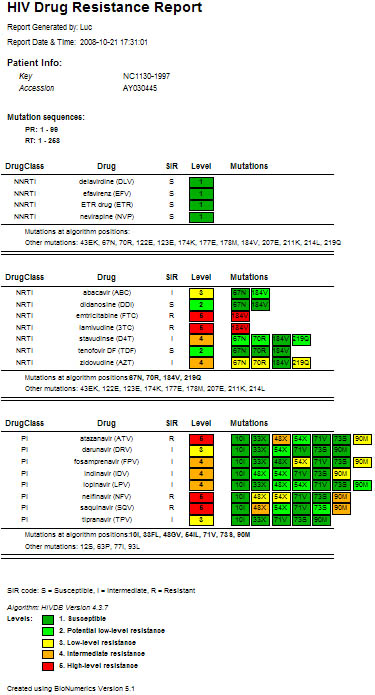
Figure 5: Detailed HIV strain resistance pattern comparison between multiple algorithms. Click on a drug to display the rule evaluation for a particular algorithm in the decision network.