In recent years, Matrix-Assisted Laser Desorption Ionization Time Of Flight Mass Spectrometry (MALDI TOF MS) has become a valuable tool for the routine identification of bacteria and fungi. It is a rapid and cost-effective method, which makes it very suitable for high-throughput applications such as pathogen identification in clinical laboratories, quality control in food industry or biomaterial validation in culture collections.
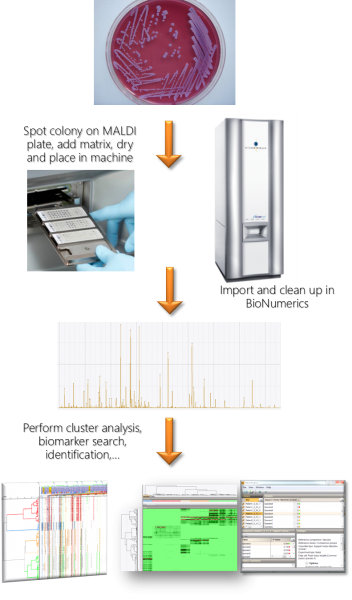
MALDI spectrum processing in BIONUMERICS
The BIONUMERICS software offers much flexibility in importing and processing spectral data. Data formats from different hardware manufacturers (e.g. Bruker, Shimadzu, VITEK MS) are supported and can be imported in the same database. The software cleans up spectra with predefined or custom processing templates. These templates can be easily adapted to any situation and stored for future use. The concept of summary spectra allows the user to summarize several technical or biological replicates into single spectra according to user-adaptable quality criteria.
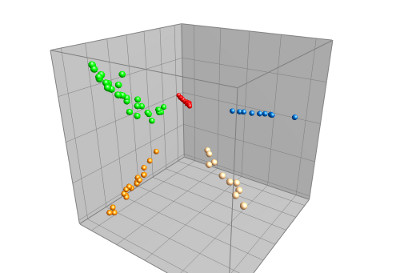
MALDI TOF spectra (either summary spectra or single spectra) can be analyzed in the Comparison window with e.g. similarity-based cluster analysis, Principal Components Analysis (PCA) or Multi Dimensional Scaling (MDS). Peak matching allows you to filter out common background peaks and to focus on a set of discriminative peaks, drastically improving the sensitivity of the analysis. The matrix mining tools are very useful for identifying potential biomarkers.
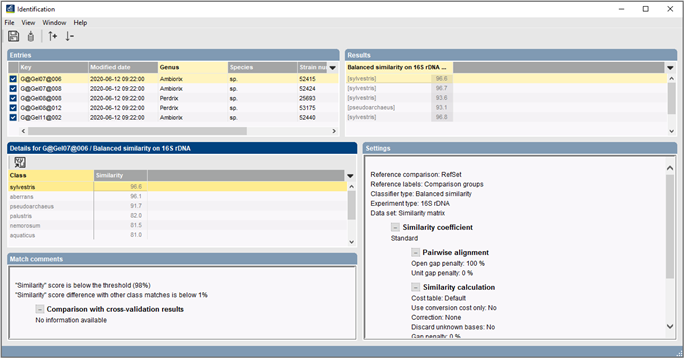
Build up your own mass spectrum database for identification
Commercially available reference databases for MALDI-TOF bacterial identification often only target clinically relevant species and come with a limited set of built-in identification methods. The BIONUMERICS software allows users to build up their own databases, based on mass spectra from organisms of their choice and store these reference sets as identification projects. Identification of unknown isolates can then be performed based on complete spectra or custom peak classes and using sophisticated classifiers such as Support Vector Machines and Naive Bayesian classifiers.