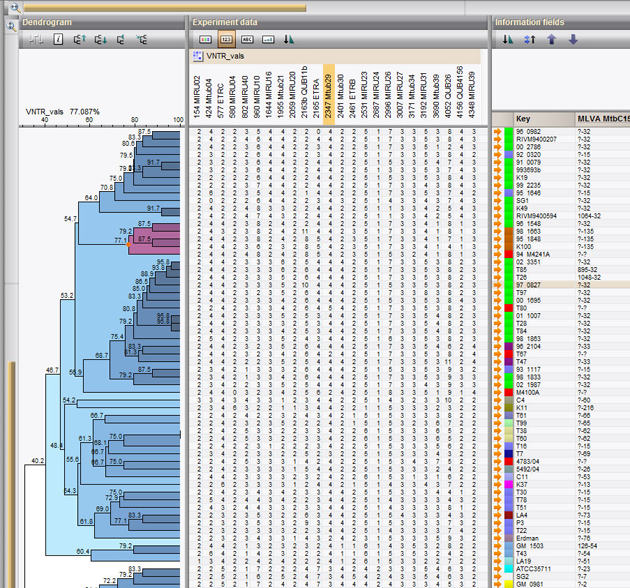
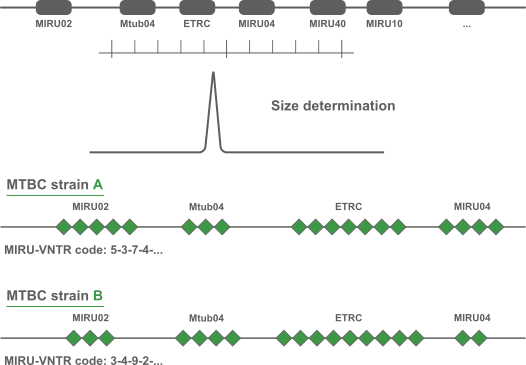
The acronym MIRU is used to denote a Mycobacterium tuberculosis-specific multiple locus VNTR (Variable Number of Tandem Repeats) analysis (MLVA) scheme. The technical aspects and analysis methods of MIRU are the same as for other MLVA schemes. In BIONUMERICS, the MIRU-VNTR plugin is based on the successful MLVA plugin.
MIRU-specific additions in the MIRU-VNTR plugin
 In addition to the automated import and analysis tools for VNTR's, the MIRU-VNTR plugin provides specific functionality for Mycobacterium interspersed repetitive units typing. The plugin automatically interprets and analyzes MIRU-VNTR data and interacts with the MIRU-VNTRplus website for typing and global epidemiology of mycobacteria.
In addition to the automated import and analysis tools for VNTR's, the MIRU-VNTR plugin provides specific functionality for Mycobacterium interspersed repetitive units typing. The plugin automatically interprets and analyzes MIRU-VNTR data and interacts with the MIRU-VNTRplus website for typing and global epidemiology of mycobacteria.
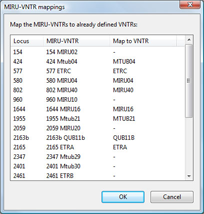
The synchronization with the MIRU-VNTRplus server allows types to be assigned automatically and new types to be submitted. The plugin optionally resolves the "MtbC 15" types, "MtbC 9" types and as a combination of both, the "MtbC 15-9" types as well.