Foodborne bacteria like Escherichia coli pose a major threat to public health. To prevent widespread infections due to these bacteria, as well as to detect outbreaks, rapid and accurate identification and characterization of these bacteria are of key importance. Conventional methods for serotyping, virulence profiling, and antimicrobial susceptibility testing are time-consuming and often require complex workflows. Alternatively, whole genome sequence data provides highly detailed genotypic information and could have the potential to replace some of these conventional methods.
E. coli genotyping in BIONUMERICS
Various E. coli genotyping tools have been bundled in one single BIONUMERICS application, making it a very user-friendly and easy-to-use typing tool. Screening for all or a subset of traits can be launched with one single click and the genotyping analysis results are summarized in an easy accessible and customizable report. The typing results are stored in the BIONUMERICS database and can be analyzed in combination with other data sets at any time.
Impressive list of genotyping tools
The E. coli genotyping application contains public databases for serotype, virulence and resistance prediction, as well as plasmid and prophage detection. The reference databases are based on the data sets available from Center for Genomic Epidemiology, DTU, combined with private knowledge. An in-silico PCR tool is also implemented, making it possible to detect e.g. Shiga toxin gene subtypes and virulence genes, mimicking the wet lab PCR.
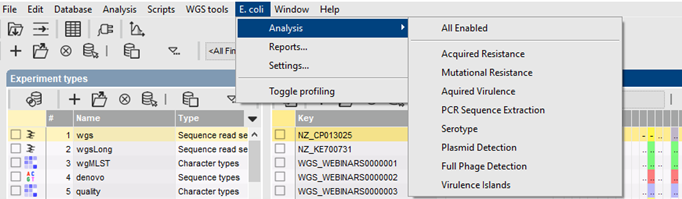
Fast BLAST screening
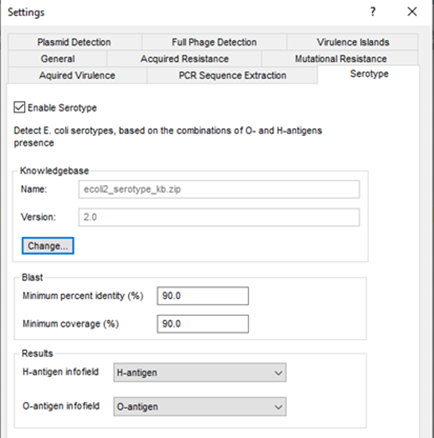
The various E. coli genotyping tools start from the assembled sequences stored in the BIONUMERICS database, and use a blast-based approach to detect and identify the genes of interest. A minimum sequence identity and minimum length coverage needs to be specified to detect the best matching target sequence in the genome.
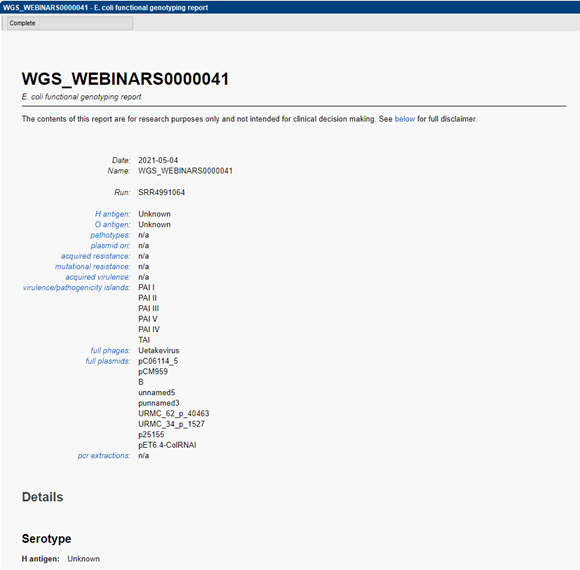
Detailed reporting
A customizable report allows to easily consult the metadata present in the linked database information fields next to the obtained genotyping results. A summary of predicted serotype, virulence and resistance prediction, as well as plasmid and prophage detection is displayed. Hyperlinks are present linking to the detailed BLAST results of the predicted traits. Detailed BLAST results include locus identifiers, BLAST similarity scores, and descriptive information on the detected genes.
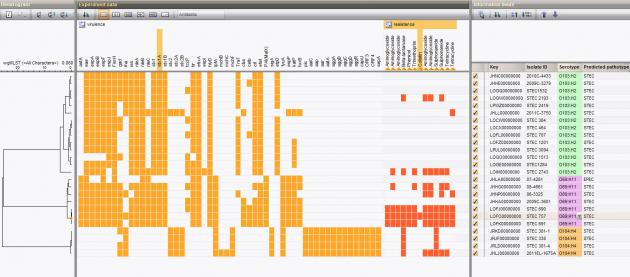
Rich database environment
The typing results are recorded as database information fields and/or character data in the BIONUMERICS database and can be clustered and analyzed together with other data sets leading to a more efficient characterization and interpretation of the data.