Classifiers are the algorithms that perform identification or classification, i.e. the process of assigning biological samples to classes (categories) based on a training set of which the class membership is already known. The samples are analyzed based on experimental data, which can be quantitative, binary or categorical.
BIONUMERICS offers a number of similarity-based classifiers, using either basic similarity values (average, maximum and minimum similarity) or more sophisticated similarity measures, such as k-Nearest Neighbors, Balanced similarity and Weighted average similarity. In addition, state-of-the-art classifiers such as Centroid distance, ANOVA-weighted Centroid distance, Shrunken Centroids, Support Vector Machines (SVMs with linear or RBF kernels) and Naive Bayesian Classifiers can be used for quick and accurate identification of complex groupings. The latter classifiers all have in common that they need to be trained before they can be used for classification of unknowns.
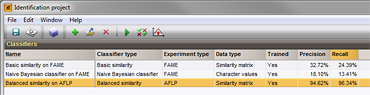 Several classifiers can be added to an identification project, allowing to side-by-side evaluation of the classifier performances.
Several classifiers can be added to an identification project, allowing to side-by-side evaluation of the classifier performances.
Cross-validation framework
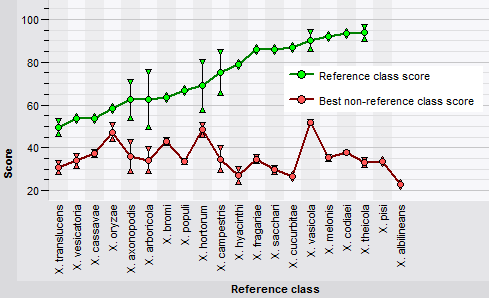
In a cross-validation analysis, the training set is split into k parts, then k-1 of these parts are used for training and the remaining part for testing. This step is repeated k times, each time using a different part for testing and the remaining parts for training. Whenever an class member is used for testing a classifier, it has not been used for training that classifier. The outcome is a so-called confusion matrix, which provides an objective estimate of the classifier’s performance.
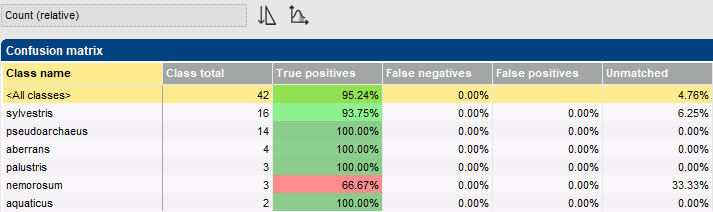
The classifier cross-validation framework in BIONUMERICS offers several pre-defined templates to conveniently generate often-used charts.
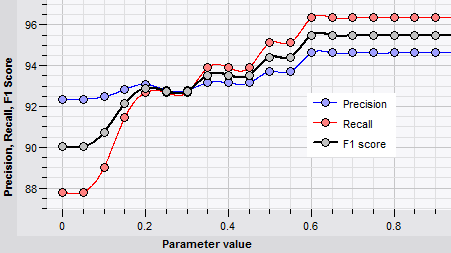
Automatic parameter optimization
As each classifiers has its own specific set of parameters, setting the optimal values can pose a challenge. BIONUMERICS helps the user to select the optimal values for algorithm parameters.