BIONUMERICS applications
BIONUMERICS is a very comprehensive software package, which has many applications in different research fields of the biological sciences. The listing below is by no means exhaustive, but gives an idea about what our software is capable of.
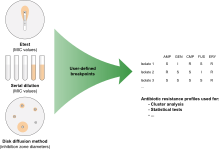
Antimicrobial susceptibility is expressed either as a Minimum Inhibitory Concentration (MIC), determined via serial dilution or Etest, or as an inhibition zone diameter when determined via the disk diffusion method. Using the Antibiotics susceptibility plugin in BIONUMERICS, cut-off values can be set and antibiotic resistance data automatically converted to SIR categories (Susceptible, Intermediate, Resistant), which can then be used for calculating dendrograms and to perform statistical analyses on.
Inter-Simple Sequence Repeat Polymerase Chain Reaction (ISSR-PCR) is a reproducible and relatively inexpensive DNA fingerprinting method for animals, plants, fungi and bacteria. It is based on the amplification of genomic regions between microsatellite loci. Running this versatile typing method on an automated DNA sizing system such as the QIAxcel Advanced System reduces the number of manipulations required as well as the time to result. Raw data files from the QIAxcel Advanced System ScreenGel® software can be imported as densitometric curve files or peak table files in BIONUMERICS with the QIAxcel plugin.
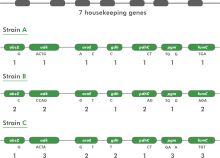
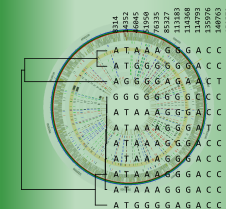
Multilocus sequence typing (MLST) is a technique whereby a number of housekeeping genes (loci) are sequenced, usually in part. In a typical MLST approach, recombination is expected to occur with a much higher frequency than point mutations. Therefore, one does not look at the total sequence similarity between strains. Instead, each sequence for a given locus is screened for identity with already known sequences for that locus. If the sequence is different, it is considered to be a new allele and is assigned a unique (arbitrary) allele number.
Since the early 90's, an increasing number of foodborne outbreaks has been investigated by national health services worldwide. Typing of all isolates in real-time became impossible, and it was decided to decentralize typing activities of bacterial and viral pathogens. Nowadays, worldwide molecular surveillance networks for bacterial (Salmonella, E.
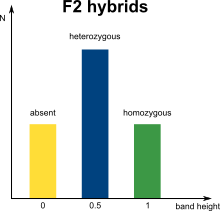
In marker assisted breeding, successful cross-over is assisted and facilitated by genetic markers. A commonly used DNA marker technology is Amplified Fragment Length Polymorphism (AFLP) analysis, where selectively amplified whole genome restriction fragments are separated via electrophoresis into a complex band profile. The occurrence of specific bands can be linked to the presence of particular phenotypic traits. In diploid organisms however, a phenotypic trait can be homozygous or heterozygous, which in theory will result in marker bands that can have double or single intensities.
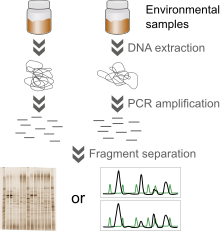
Bacterial community fingerprinting refers to a number of cultivation-independent methods that are used to measure biodiversity in environmental samples or to profile microbial community structures, e.g. in response to experimental conditions. Examples include Denaturing Gradient Gel Electrophoresis (DGGE), Temperature Gradient Gel Electrophoresis (TGGE), Terminal Restriction Fragment Length Polymorphism (T-RFLP) and (Automated) Ribosomal Intergenic Spacer Analysis (ARISA).
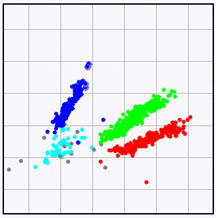
BIONUMERICS features an analysis suite that allows you to detect Single Nucleotide Polymorphisms (SNPs) on whole genomes sequences and perform cluster analyses on the resulting whole genome SNP (wgSNP) matrix.
The BIONUMERICS SARS-CoV-2 plugin tool facilitates the processing and analysis of SARS-CoV-2 genomic sequences, whether downloaded from a public data repository or generated locally.
The BIONUMERICS HIV drug resistance analysis application is a plugin-based expert system for automated prediction of antiretroviral drug resistance/susceptibility of HIV strains based on pol gene sequencing.
The TaqMan® SNP genotyping technology utilizes the 5’ nuclease activity of Taq polymerase to generate a fluorescent signal during PCR. For each SNP, the assay uses two TaqMan® probes that differ in sequence only at the SNP site, with one probe complementary to the wild-type allele and the other to the variant allele. The technique utilizes the FRET technology whereby a 5’ reporter dye and a 3’ quencher dye are covalently linked to the wild-type and variant allele probes.
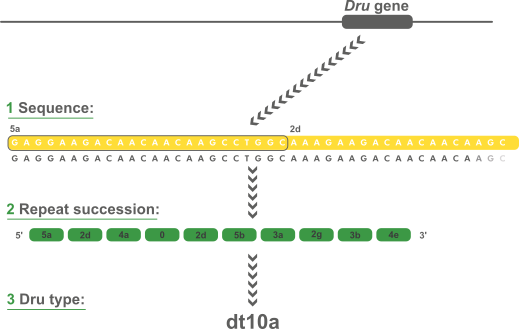
Variable-Number Tandem Repeats (VNTR's) are well known for their high mutation rate and are therefore widely used for subtyping. Some VNTR's, however, exhibit polymorphism in their individual repeat sequences. By sequencing the polymorphic repeat region, each new repeat variant determined can be assigned a unique repeat code. The repeat succession for a given strain in turn, determines that strain's VNTR type.
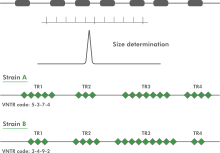
Multi Locus VNTR Analysis (MLVA) is a molecular typing method to subtype microbial isolates based upon the Variable copy Numbers of Tandem Repeats (VNTR). A VNTR typically exhibits a large range of copy numbers, even among highly related bacterial strains. For a selected set of tandem repeats, copy number analysis reveals insights about the relationships at a micro-evolutionary level.
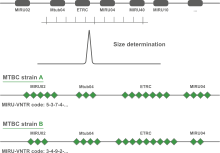
The acronym MIRU is used to denote a Mycobacterium tuberculosis-specific multiple locus VNTR (Variable Number of Tandem Repeats) analysis (MLVA) scheme. The technical aspects and analysis methods of MIRU are the same as for other MLVA schemes. In BIONUMERICS, the MIRU-VNTR plugin is based on the successful MLVA plugin.
In recent years, Matrix-Assisted Laser Desorption Ionization Time Of Flight Mass Spectrometry (MALDI TOF MS) has become a valuable tool for the routine identification of bacteria and fungi. It is a rapid and cost-effective method, which makes it very suitable for high-throughput applications such as pathogen identification in clinical laboratories, quality control in food industry or biomaterial validation in culture collections.
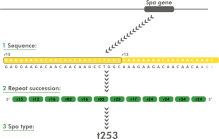
The spa typing technique uses the sequence of a polymorphic VNTR in the 3' coding region of the S. aureus-specific staphylococcal protein A (spa). Each new base composition of the polymorphic repeat found in a strain is assigned a unique repeat code. The repeat succession for a given strain determines its spa type.
Various E. coli genotyping tools have been bundled in one single BIONUMERICS application, making it a very user-friendly and easy-to-use typing tool. Screening for all or a subset of traits can be launched with one single click and the genotyping analysis results are summarized in an easy accessible and customizable report. The typing results are stored in the BIONUMERICS database and can be analyzed in combination with other data sets at any time.
As next-generation sequencing, which offers a fast and cost-effective way to sequence bacterial genomes, is increasingly replacing Sanger sequencing, conventional MLST can be extended to whole genome MLST (wgMLST). Since many more loci (typically 1500 – 4000) are considered in wgMLST, a much higher typing resolution can be obtained.
BIONUMERICS software offers an integrated platform for the analysis of PFGE fingerprints. Store all your epidemiological info and gel images in one database. With powerful statistical tools, BIONUMERICS makes it easy to perform cluster analysis and calculate dendrograms.